.png?width=1440&name=Julia%20header%20(1).png)
Sep 20, 2023
RUNNING JULIA ON GRAPHCORE IPUS
Written By:
Mosè Giordano
.png?width=1440&name=Julia%20header%20(1).png)
Sep 20, 2023
Written By:
Mosè Giordano
We're Hiring
Join us and build the next generation AI stack - including silicon, hardware and software - the worldwide standard for AI compute
Join our teamI am a Research Software Developer at University College London and user of the Julia programming language since 2016. I am also curious about trying out Julia on interesing hardware - which led me to implementing it on the Graphcore IPUs. However, I am not a compiler engineer.
You can try running code written in Julia on the IPU for free using a number of sample notebooks on Paperspace Gradient.
You may also like to watch a presentation I gave on implementing Julia on the Graphcore IPU at JuliaCon 2023.
Julia is a modern, dynamic, general-purpose, compiled programming language. It's interactive ("like Python"), can be used in a REPL or notebooks, like Jupyter (it's the "Ju") or Pluto (this one🎈). Julia has a runtime which includes a just-in-time (JIT) compiler and a garbage collector (GC), for automatic memory management.
Julia is mainly used for numerical computing, diffential equations solvers suite is quite popular.
Main paradigm of Julia is multiple dispatch, what functions do depend on type and number of all arguments.

From "My Target Audience" by Matthijs Cox
We developed a package called IPUToolkit.jl to interface the Poplar SDK:
using IPUToolkit.Poplar
let
# Set up graph and program
device = Poplar.get_ipu_device()
target = Poplar.DeviceGetTarget(device)
graph = Poplar.Graph(target)
prog = Poplar.ProgramSequence()
# Create CPU array
h3 = zeros(Float32, 4, 4)
# Create IPU tensors
c1 = Poplar.GraphAddConstant(graph, Float32[1.0, 1.5, 2.0, 2.5])
v1 = similar(graph, c1, "v1")
v2 = similar(graph, c1, "v2")
v3 = similar(graph, h3, "v3")
v4 = Poplar.GraphAddVariable(graph, Poplar.INT(), UInt64[10], "v4")
# Tensors tile mapping
Poplar.GraphSetTileMapping(graph, v1, 0)
for i in UInt64(0):UInt64(3)
Poplar.GraphSetTileMapping(graph, v2[i], i)
end
Poplar.GraphSetTileMapping(graph, v3, 0)
Poplar.GraphSetTileMapping(graph, v4, 0)
Poplar.GraphSetTileMapping(graph, c1, 0)
# Copy `c1` to `v1` and print `v1`
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramCopy(c1, v1))
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramPrintTensor("v1-debug", v1))
# Copy `v1` to `v2` and print `v2` (should be same as `v1` above)
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramCopy(v1, v2))
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramPrintTensor("v2-debug", v2))
# Prepare copying data between CPU and IPU
Poplar.GraphCreateHostWrite(graph, "v3-write", v3)
Poplar.GraphCreateHostRead(graph, "v3-read", v3)
v1slice = Poplar.TensorSlice(v1, 0, 3)
v3slice = Poplar.TensorSlice(v3, UInt64[1, 1], UInt64[2, 4])
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramCopy(v1slice, v3slice))
# Read three batches of 10 `Int32`s from the strea `inStream` into `v4` and print values
inStream = Poplar.GraphAddHostToDeviceFIFO(graph, "v4-input-stream", Poplar.INT(), 10)
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramCopy(inStream, v4))
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramPrintTensor("v4-0", v4))
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramCopy(inStream, v4))
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramPrintTensor("v4-1", v4))
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramCopy(inStream, v4))
Poplar.ProgramSequenceAdd(prog, Poplar.ProgramPrintTensor("v4-2", v4))
flags = Poplar.OptionFlags()
Poplar.OptionFlagsSet(flags, "debug.instrument", "true")
engine = Poplar.Engine(graph, prog, flags)
Poplar.EngineLoad(engine, device)
Poplar.EngineWriteTensor(engine, "v3-write", h3)
# Create data to stream to `v4`
inData = Int32.(1:30)
# Connect data to stream
Poplar.EngineConnectStream(engine, "v4-input-stream", inData)
# Run the engine
Poplar.EngineRun(engine, 0)
# Write IPU tensor `v3` to CPU array `h3`
Poplar.EngineReadTensor(engine, "v3-read", h3)
# Release device
Poplar.detach_devices()
# Print value of CPU array `h3`
print("h3 data: ")
display(h3')
end
Trying to attach to device 0...
Successfully attached to device 0
v1-debug: [1.0000000 1.5000000 2.0000000 2.5000000]
v2-debug: [1.0000000 1.5000000 2.0000000 2.5000000]
v4-0: [ 1 2 3 4 5 6 7 8 9 10]
v4-1: [11 12 13 14 15 16 17 18 19 20]
v4-2: [21 22 23 24 25 26 27 28 29 30]
h3 data: 4×4 adjoint(::Matrix{Float32}) with eltype Float32:
0.0 0.0 0.0 0.0
0.0 1.0 1.5 2.0
0.0 0.0 0.0 0.0
0.0 0.0 0.0 0.0
But we are just writing C++ code in Julia, nothing excitingly new... Can we do more?
.png?width=902&height=370&name=LysTgpt%20(1).png)
(A high-level diagram of a compiler pipeline, with emphasis on the front-end, from "What can we learn from how compilers are designed?")
The only existing implementation of Julia is based on the LLVM compiler, a popular modular compilation framework.
With Julia we can easily inspect each stage of the compilation pipeline.
Meta.@dump 1.0 + 2.0
Expr
head: Symbol call
args: Array{Any}((3,))
1: Symbol +
2: Float64 1.0
3: Float64 2.0
CodeInfo(
1 ─ %1 = Base.add_float(x, y)::Float64
└── return %1
) =>Float64)
@code_typed 1.0 + 2.0
@code_llvm debuginfo=:none 1.0 + 2.0
define double @"julia_+_4759"(double %0, double %1) #0 {
top:
%2 = fadd double %0, %1
ret double %2
}
@code_native debuginfo=:none 1.0 + 2.0
.text
.file "+"
.globl "julia_+_4795" # -- Begin function julia_+_4795
.p2align 4, 0x90
.type "julia_+_4795",@function
"julia_+_4795": # @"julia_+_4795"
# %bb.0: # %top
push rbp
mov rbp, rsp
vaddsd xmm0, xmm0, xmm1
pop rbp
ret
.Lfunc_end0:
.size "julia_+_4795", .Lfunc_end0-"julia_+_4795"
# -- End function
.section ".note.GNU-stack","",@progbits
For more details on the Julia compiler watch the talk "Compiling Julia: Making dynamic programs run fast" by Valentin Churavy.
It turns out also the Poplar compiler, the compiler developed by Graphcore for generating the native code for the IPU, is based on LLVM. When you compile a codelet for the IPU you, the Poplar compiler performs the same pipeline we have seen above.
This means that Julia and the Poplar compiler can actually talk the same language: the LLVM IR.
We added to IPUToolkit.jl the capability to generate code for the IPU with Julia. All in all, this package has the following goals:
.png?width=1103&height=610&name=LLVM%20(1).png)
We can use the IPUToolkit.jl package to write IPU codelets in Julia, generating LLVM IR code which is then compiled to native code with the Poplar compiler. The code generation via LLVM is based on the GPUCompiler.jl package, which is a generic framework for generating LLVM IR code for specialised targets, not limited to GPUs despite the historical name.
The code inside a codelet has the same limitations as all the compilation models based on GPUCompiler.jl:
The LLVM IR is mostly target-independent, but not completely. Also, some details, like width of vectorisation registers, can be better tailored for the target if its properties are known correctly. For most of this project I actually generated LLVM IR for the host CPU, which "works", but is not optimal.
I recently managed to link Julia to Graphcore's fork of LLVM, which allowed me to directly generate code for the IPU (the "Colossus" target), however this combination is highly experimental and causes some unexpected errors.
Inspired by Owain Kenway's pi_examples in several different languages.
using IPUToolkit.IPUCompiler
# Disable spinner to show progress of codelet compilation,
# Pluto notebooks have their own progress indicator.
IPUCompiler.PROGRESS_SPINNER[] = false;
# Do not automatically delete LLVM modules of codelets.
IPUCompiler.KEEP_LLVM_FILES[] = true;
# Limit run-time of codelets
ENV["POPLAR_RUNTIME_OPTIONS"] = """{"target.hostSyncTimeout":"60"}""";
let
device = Poplar.get_ipu_device()
target = Poplar.DeviceGetTarget(device)
graph = Poplar.Graph(target)
num_tiles = Int(Poplar.TargetGetNumTiles(target))
n::UInt32 = typemax(UInt32) ÷ num_tiles
unroll::UInt32 = 1
iszero(rem(n, unroll)) || error("n ($(n)) is not an integer multiple of unroll factor ($(unroll))")
num_steps::UInt32 = num_tiles * n
slice::Float32 = 1 / num_steps
tile_clock_frequency = Poplar.TargetGetTileClockFrequency(target)
ids = collect(UInt32.(0:(num_tiles - 1)))
sums = similar(ids, Float32)
cycles = similar(ids)
# With `@eval` we can interpolate (using `$`) global variables
# inside the kernel.
@eval function pi_kernel(i::T) where {T<:Integer}
sum = 0f0
for j in (i * $(n)):$(unroll):((i + one(T)) * $(n) - one(T))
Base.Cartesian.@nexprs $(Int64(unroll)) idx -> begin
x = (j + Float32(idx - 1) - 0.5f-1) * $(slice)
sum += 4 / (1 + x ^ 2)
end
end
return sum
end
@codelet graph function Pi(in::VertexVector{UInt32, In},
out::VertexVector{Float32, Out},
cycles::VertexVector{UInt32, Out})
cycles[begin] = @ipuelapsed(out[begin] = pi_kernel(in[begin]))
end
# Type and shape of tensors automatically inferred from input arrays
ids_ipu = Poplar.GraphAddConstant(graph, ids)
sums_ipu = similar(graph, sums, Float32, "sums");
cycles_ipu = similar(graph, cycles, UInt32, "cycles");
prog = Poplar.ProgramSequence()
# Automatically add the codelet defined above to the program
# and spread it across all the tiles
add_vertex(graph, prog, 0:(num_tiles - 1), Pi, ids_ipu, sums_ipu, cycles_ipu)
# Simplified API for `GraphCreateHostRead`, we can use fewer arguments
Poplar.GraphCreateHostRead(graph, "sums-read", sums_ipu)
Poplar.GraphCreateHostRead(graph, "cycles-read", cycles_ipu)
engine = Poplar.Engine(graph, prog)
Poplar.EngineLoadAndRun(engine, device)
# Simplified API also for `EngineReadTensor`
Poplar.EngineReadTensor(engine, "sums-read", sums)
Poplar.EngineReadTensor(engine, "cycles-read", cycles)
Poplar.detach_devices()
pi = sum(sums) * slice
time = round(maximum(cycles) / tile_clock_frequency; sigdigits=4)
print("""
Calculating PI using:
$(num_steps) slices
$(num_tiles) IPU tiles
loop unroll factor $(Int(unroll))
Obtained value of PI: $(pi)
Time taken: $(time) seconds ($(maximum(cycles)) cycles at $(round(tile_clock_frequency / 1e9; sigdigits=3)) GHz)
""")
end;
Trying to attach to device 0...
Successfully attached to device 0
Calculating PI using:
4294966272 slices
1472 IPU tiles
loop unroll factor 1
Obtained value of PI: 3.1499734
Time taken: 0.123 seconds (227586882 cycles at 1.85 GHz)
We can write an equivalent C++ program to compare performance, to check how Julia code generation is faring. This is the codelet:
#include <poplar/Vertex.hpp>
using namespace poplar;
class VertexPi : public Vertex {
public:
Input<unsigned> id;
Input<unsigned> n;
Input<float> slice;
Output<Vector<float>> out;
Output<Vector<unsigned>> cycles;
bool compute() {
float sum = 0;
unsigned start = __builtin_ipu_get_scount_l();
for (unsigned j=id * n; j < (id + 1) * n; j++) {
float x = (j - 0.5f) * slice;
sum += 4.0f / (1.0f + x * x);
}
unsigned end = __builtin_ipu_get_scount_l();
out[0] = sum;
cycles[0] = end - start;
return true;
}
};
run(`$(cpp_exe_path)`);
Using HW device ID: 0
Calculating PI using:
4294966272 slices
1472 IPU tiles
Obtained value of PI: 0.146298
Time taken: 0.141946 seconds (262599924 cycles at 1.85 GHz)
Main performance difference between the C++ and the Julia codelets in this case is due to loop unrolling. Some experimentations showed that when both code have loops similarly unrolled then performance is perfectly comparable. This showed that Julia can be a valid front-end for writing code for the IPU.
IPUToolkit.jl provides some macros for quickly benchmarking parts of a codelet: @ipucycles, @ipushowcycles, @ipuelapsed (the latter was already used above).
let
device = Poplar.get_ipu_device()
target = Poplar.DeviceGetTarget(device)
graph = Poplar.Graph(target)
@codelet graph function Benchmark(input::VertexVector{Float32, In},
output::VertexVector{Float32, Out})
for idx in eachindex(input, output)
x = input[idx]
@ipushow x
x = @ipushowcycles sin(x)
x = @ipushowcycles cos(x)
x = @ipushowcycles exp(x)
output[idx] = x
end
end
in_ipu = Poplar.GraphAddConstant(graph, randn(Float32, 5))
out_ipu = similar(graph, in_ipu);
prog = Poplar.ProgramSequence()
add_vertex(graph, prog, Benchmark, in_ipu, out_ipu)
engine = Poplar.Engine(graph, prog)
Poplar.EngineLoadAndRun(engine, device)
Poplar.detach_devices()
end
Trying to attach to device 0...
Successfully attached to device 0
T[0.5]: x = 0.433483
T[0.5]: sin(x):
T[0.5]: 14418 cycles
T[0.5]: cos(x):
T[0.5]: 13320 cycles
T[0.5]: exp(x):
T[0.5]: 18 cycles
T[0.5]: x = -1.012771
T[0.5]: sin(x):
T[0.5]: 14448 cycles
T[0.5]: cos(x):
T[0.5]: 15834 cycles
T[0.5]: exp(x):
T[0.5]: 18 cycles
T[0.5]: x = 0.436158
T[0.5]: sin(x):
T[0.5]: 14430 cycles
T[0.5]: cos(x):
T[0.5]: 13302 cycles
T[0.5]: exp(x):
T[0.5]: 18 cycles
T[0.5]: x = 1.835203
T[0.5]: sin(x):
T[0.5]: 14562 cycles
T[0.5]: cos(x):
T[0.5]: 15798 cycles
T[0.5]: exp(x):
T[0.5]: 18 cycles
T[0.5]: x = -0.725937
T[0.5]: sin(x):
T[0.5]: 14592 cycles
T[0.5]: cos(x):
T[0.5]: 13308 cycles
T[0.5]: exp(x):
T[0.5]: 18 cycles
Note: This isn't a replacement for profiling, which is still an invaluable tool (but JIT complicats stacktraces), however these macros can be useful for quick feedback.
Cycle-count-based benchmarking is not reliable for blocks taking longer than typemax(UInt32) = 4294967295 cycles, about 2-3 seconds depending on your IPU model. You need to have a feeling whether the block you want to benchmark would overflow the counter.
We can also use third-party packages inside codelets, as long as the requirements mentioned above are met: no memory allocations, fully inferrable code. The StaticArrays.jl allows you to create arrays on the stack and do basic linear algebra operations with them. Code involving static arrays is also usally easy to infer for the compiler.
StaticArrays.jl isn't more efficient than using specialised Popops linear algebra routines, but nonetheless is a good example of usage of external packages inside IPU codelets written in Julia.
using StaticArrays
let
device = Poplar.get_ipu_device()
target = Poplar.DeviceGetTarget(device)
graph = Poplar.Graph(target)
N = 16
mat1 = randn(Float32, N)
mat2 = randn(Float32, N)
mul = PoplarVector{Float32}(undef, N)
inverse = PoplarVector{Float32}(undef, N)
@ipuprogram device begin
function MatMul(in1::VertexVector{Float32, In},
in2::VertexVector{Float32, In},
mul::VertexVector{Float32, Out})
m1 = @inbounds SMatrix{4,4,Float32,16}(in1)
m2 = @inbounds SMatrix{4,4,Float32,16}(in2)
mul .= (m1 * m2)[:]
end
function Inverse(input::VertexVector{Float32, In},
inverse::VertexVector{Float32, Out})
m = @inbounds SMatrix{4,4,Float32,16}(input)
inverse .= inv(m)[:]
end
MatMul(mat1, mat2, mul)
Inverse(mul, inverse)
jl_mul = mul
jl_inv = inverse
end
Poplar.detach_devices()
jl_mul = reshape(jl_mul, 4, 4)
@show reshape(mat1, 4, 4) * reshape(mat2, 4, 4) ≈ jl_mul
@show reshape(jl_inv, 4, 4) ≈ inv(jl_mul)
end;
Trying to attach to device 0...
Successfully attached to device 0
reshape(mat1, 4, 4) * reshape(mat2, 4, 4) ≈ jl_mul = true
reshape(jl_inv, 4, 4) ≈ inv(jl_mul) = true
Real numbers constitue a continuous set
Stochastic rounding is an alternative rounding mode to classic deterministic rounding, which randomly rounds a number
 Common choices are
Common choices are
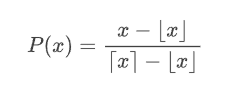 In the following we'll always talk about the latter probability function
In the following we'll always talk about the latter probability function
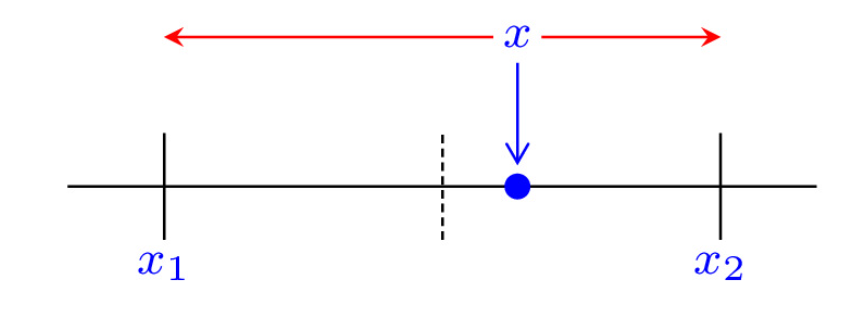 (source: "What Is Stochastic Rounding?" by Nick Higham)
(source: "What Is Stochastic Rounding?" by Nick Higham)
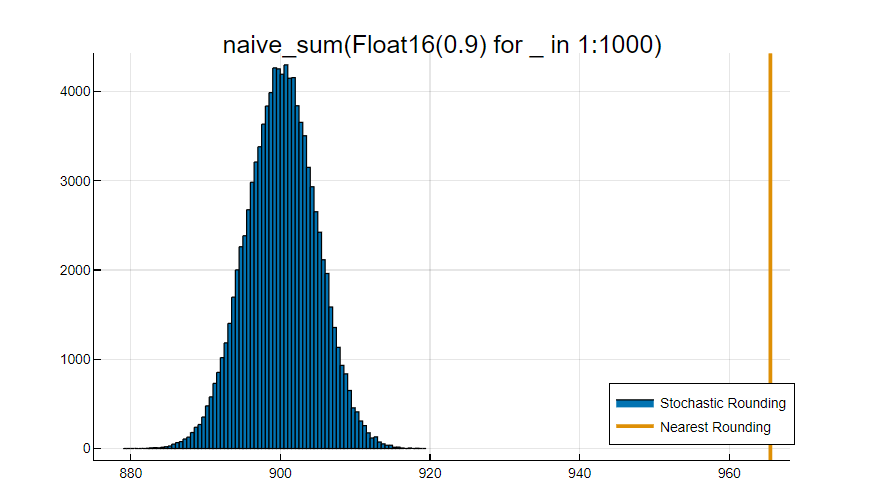
Stochastic rounding is useful because the average result of operations matches the expected mathematical result. In a statistical sense, it retains some of the information that is discarded by a deterministic rounding scheme, smoothing out numerical rounding errors due to limited precisions. This is particularly important when using low-precision floating point numbers like Float16. For contrast, deterministic rounding modes like nearest rounding introduce a bias, which is more severe as the precision of the numbers is lower.
The IPU is one of the very few processors available with hardware support for stochastic rounding.
Let's do an exercise on the CPU with classical nearest rounding. We define a function to do the naive sequential sum of a vector of numbers, because the sum function in Julia uses pairwise summation, which would have better accuracy.
naive_sum(v) = foldl(+, v)
x_sr = fill(Float16(0.9), 1000);
naive_sum(x_sr)
naive_sum(x_sr) ≈ x_sr[1] * length(x_sr)
Now let's write an IPU program which computes the sum of x_sr multiple times with stochastic rounding:
sums_sr = let
device = Poplar.get_ipu_device()
target = Poplar.DeviceGetTarget(device)
graph = Poplar.Graph(target)
prog = Poplar.ProgramSequence()
Poplar.PoplarSetStochasticRounding(graph, prog, true, "")
@codelet graph function stochastic_rounding(x::VertexVector{Float16, In}, sums::VertexVector{Float16, Out})
for idx in eachindex(sums)
sums[idx] = naive_sum(x)
end
end
sums_sr = Vector{Float16}(undef, 100_000)
x_ipu = Poplar.GraphAddConstant(graph, x_sr)
sums_ipu = similar(graph, sums_sr)
add_vertex(graph, prog, stochastic_rounding, x_ipu, sums_ipu)
# Prepare tensors read
Poplar.GraphCreateHostRead(graph, "sums-read", sums_ipu)
# Run the program
engine = Poplar.Engine(graph, prog)
Poplar.EngineLoadAndRun(engine, device)
# Read the output tensors back to the CPU
Poplar.EngineReadTensor(engine, "sums-read", sums_sr)
Poplar.detach_devices()
sums_sr
end;
Trying to attach to device 0...
Successfully attached to device 0
using Plots: plot, plot!, contour, surface, heatmap, histogram, vline!, plotlyjs
import PlotlyJS
plotlyjs();
using Statistics
extrema(sums_sr)
mean(Float64.(sums_sr))
std(Float64.(sums_sr))
median(sums_sr)
all(isapprox(sum(Float64.(x_sr))), sums_sr)
If we want to use external packages on the IPU, we can look for similar solutions for the GPU. If they have been made generic enough to work with different backends chances are good that they can be used on the IPU as well.
An example is DiffEqGPU.jl, a suite of differential equations solvers part of the SciML ecosystem that can be run on different kinds of GPUs (Nvidia, AMD, Intel, Metal), but it provides basic functionalities that we can reuse on the IPU as well.
using DiffEqGPU, OrdinaryDiffEq
N_lotka_volterra = 20_000;
begin
T_lotka_volterra = Float16
α_lotka_volterra_range = range(T_lotka_volterra(0.1); step=T_lotka_volterra(0.1), length=64)
β_lotka_volterra_range = range(T_lotka_volterra(0.1); step=T_lotka_volterra(0.1), length=23)
end;
# Create a range of different input parameters
lotka_volterra_parameters = let
α = repeat(α_lotka_volterra_range; inner=23)
β = repeat(β_lotka_volterra_range; outer=64)
γ = repeat([T_lotka_volterra(3)], length(β))
δ = repeat([T_lotka_volterra(1)], length(β))
zip(α, β, γ, δ) |> Iterators.flatten |> collect
end;
function lotka_volterra(u, p, t)
α, β, γ, δ = p
🐰, 🦊 = u
d🐰 = α * 🐰 - β * 🐰 * 🦊
d🦊 = -γ * 🦊 + δ * 🐰 * 🦊
return SVector{2}(d🐰, d🦊)
end;
function lotka_volterra_cpu(p::Vector,
n::Integer,
timestep::Real,
FT::DataType=Float64)
u1 = Vector{FT}(undef, n)
u2 = Vector{FT}(undef, n)
u0 = @SVector [FT(1); FT(1)]
svp = @inbounds SVector{4, FT}(p)
integ = DiffEqGPU.init(GPUTsit5(), lotka_volterra, false, u0, FT(0), FT(timestep), svp, nothing, CallbackSet(nothing), true, false)
for idx in eachindex(u1, u2)
DiffEqGPU.step!(integ, integ.t + integ.dt, integ.u)
u1[idx] = integ.u[1]
u2[idx] = integ.u[2]
end
return u1, u2
end;
u1_16, u2_16, u1_16_sr, u2_16_sr = let
device = Poplar.get_ipu_device()
target = Poplar.DeviceGetTarget(device)
graph = Poplar.Graph(target)
tile_clock_frequency = Poplar.TargetGetTileClockFrequency(target)
num_tiles = Int(Poplar.TargetGetNumTiles(target))
tiles = 0:(num_tiles - 1)
@codelet graph function solve_lv(u1::VertexVector{Float16, Out},
u2::VertexVector{Float16, Out},
p::VertexVector{Float16, In},
cycles::VertexVector{UInt32, Out})
u0 = @SVector [Float16(1); Float16(1)]
svp = @inbounds SVector{4, Float16}(p)
integ = DiffEqGPU.init(GPUTsit5(), lotka_volterra, false, u0, Float16(0), Float16(0.001), svp, nothing, CallbackSet(nothing), true, false)
cycles[begin] = @ipuelapsed for idx in eachindex(u1, u2)
DiffEqGPU.step!(integ, integ.t + integ.dt, integ.u)
u1[idx] = integ.u[1]
u2[idx] = integ.u[2]
end
end
FT = T_lotka_volterra
len = N_lotka_volterra * (length(lotka_volterra_parameters) ÷ 4)
u1_16 = Vector{FT}(undef, len)
u2_16 = Vector{FT}(undef, len)
u1_16_sr = Vector{FT}(undef, len)
u2_16_sr = Vector{FT}(undef, len)
cycles = Vector{UInt32}(undef, num_tiles)
cycles_sr = Vector{UInt32}(undef, num_tiles)
p_ipu = Poplar.GraphAddConstant(graph, lotka_volterra_parameters)
# Convenient function for defining the name of a read operation
_read(name::String, sr::Bool) = "$(name)-$(sr ? "sr-" : "")read"
for sr in (true, false)
prog = Poplar.ProgramSequence()
Poplar.PoplarSetStochasticRounding(graph, prog, sr, "")
u1_ipu = similar(graph, sr ? u1_16_sr : u1_16)
u2_ipu = similar(graph, sr ? u2_16_sr : u2_16)
cycles_ipu = similar(graph, sr ? cycles_sr : cycles)
# Run the codelet defined above on all tiles,
# with tensors evenly spread across all cores.
add_vertex(graph, prog, tiles, solve_lv, u1_ipu, u2_ipu, p_ipu, cycles_ipu)
# Prepare tensors read
Poplar.GraphCreateHostRead(graph, _read("u1", sr), u1_ipu)
Poplar.GraphCreateHostRead(graph, _read("u2", sr), u2_ipu)
Poplar.GraphCreateHostRead(graph, _read("cycles", sr), cycles_ipu)
# Run the program
engine = Poplar.Engine(graph, prog)
Poplar.EngineLoadAndRun(engine, device)
# Read the output tensors back to the CPU
Poplar.EngineReadTensor(engine, _read("u1", sr), sr ? u1_16_sr : u1_16)
Poplar.EngineReadTensor(engine, _read("u2", sr), sr ? u2_16_sr : u2_16)
Poplar.EngineReadTensor(engine, _read("cycles", sr), sr ? cycles_sr : cycles)
end
Poplar.detach_devices()
time = round(maximum(cycles) / tile_clock_frequency; sigdigits=3)
time_sr = round(maximum(cycles_sr) / tile_clock_frequency; sigdigits=3)
@info "Solving $(length(lotka_volterra_parameters) ÷ 4) ODEs on $(num_tiles) tiles took $(time) seconds with NR, and $(time_sr) with SR."
u1_16, u2_16, u1_16_sr, u2_16_sr
end;
Trying to attach to device 0...
Successfully attached to device 0
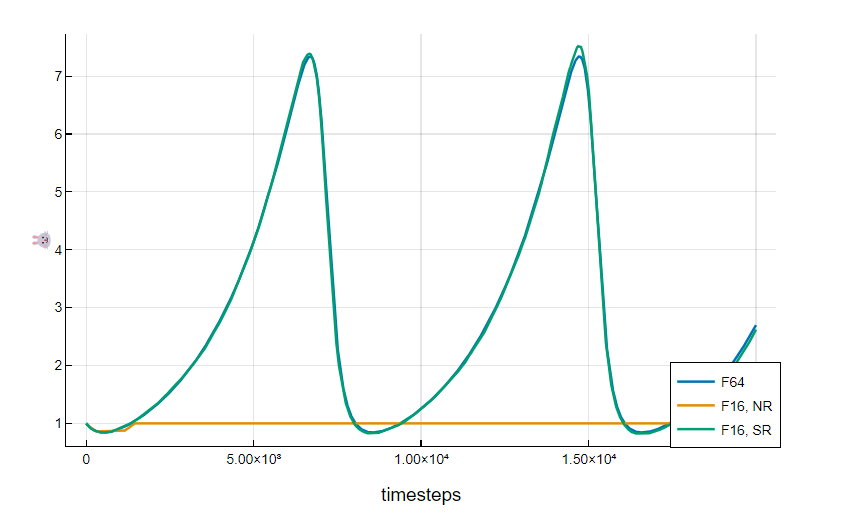
Solving 1472 ODEs on 1472 tiles took 0.0145 seconds with NR, and 0.0145 with SR.

 Automatic differentiation
Automatic differentiationIn mathematics, contrary to integration, computing the derivative of an expression is a mechanical process. Wouldn't it be nice to automatically compute the exact derivative of mathematical functions in your code?
Automatic differentiation (autodiff in short) is a set of techniques to automatically compute the derivative of a function in a computer program.
Enzyme is a source code transformation autodiff engine operating at the LLVM level: it analysis the LLVM IR of the source code and takes the derivative of all the instructions, based on the built-in differentiation rules. The fact that it works at the LLVM level means that it's mostly front-end-language-agnostic and can be used to take derivative of source code combining multiple languages or using parallelisation frameworks ("Scalable automatic differentiation of multiple parallel paradigms through compiler augmentation" won Best Student Paper award at SuperComputing 2022). Also, Enzyme generates code at compile-time, it doesn't run itself on the target system: it's the perfect candidate for use in an IPU program!
using Enzyme
Enzyme.Compiler.enzyme_code_llvm(stdout, sin, Active, Tuple{Active{Float64}})
; Function Attrs: alwaysinline nofree nosync readnone
define [1 x [1 x double]] @diffejulia_sin_10561mustwrap_inner_1wrap(double %0, double %1) #2 {
entry:
%2 = call fast double @llvm.cos.f64(double %0)
%3 = fmul fast double %2, %1
%.unpack1 = insertvalue [1 x double] zeroinitializer, double %3, 0
%4 = insertvalue [1 x [1 x double]] zeroinitializer, [1 x double] %.unpack1, 0
ret [1 x [1 x double]] %4
}
Evereyone's favourite example function for optimisation problems.
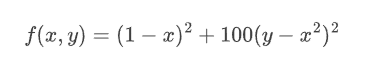
rosenbrock(x, y=4) = (1 - x) ^ 2 + 100 * (y - x ^ 2) ^ 2
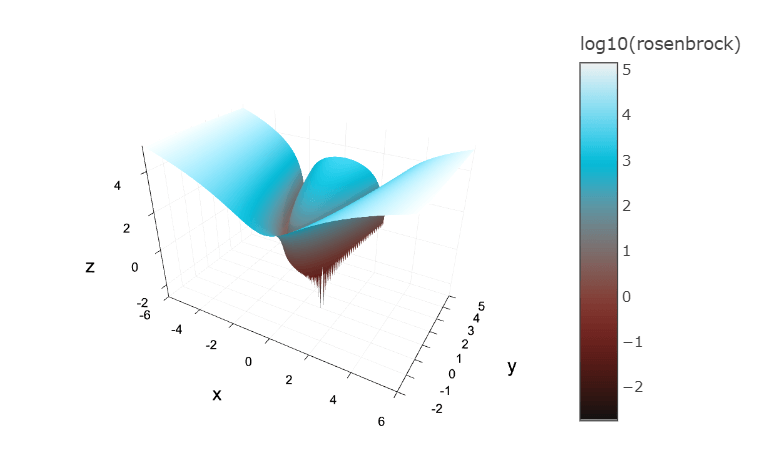
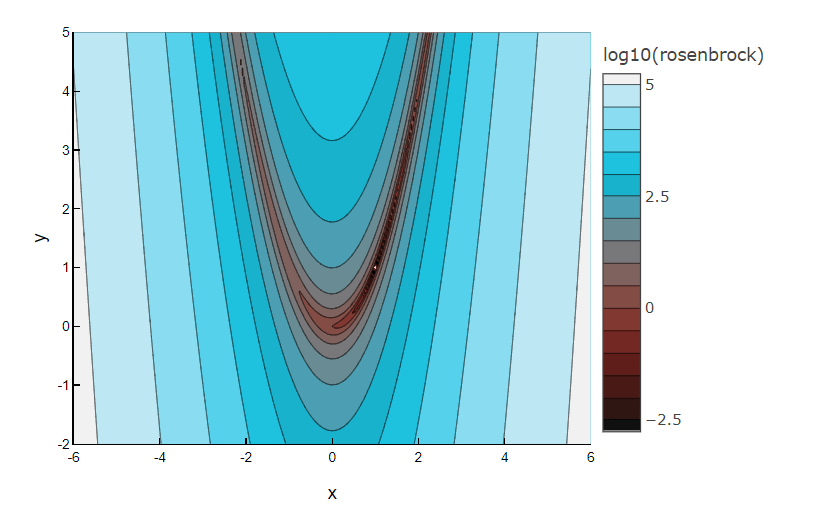 Finding the valley is trivial, but converging to the global minimum is hard.
Finding the valley is trivial, but converging to the global minimum is hard.
The Rosenbrock function is a polynomial function, easy to compute its gradient:
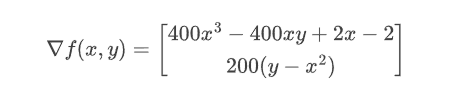 We can ask Enzyme to compute the partial derivative on the second argument:
We can ask Enzyme to compute the partial derivative on the second argument:
Enzyme.Compiler.enzyme_code_llvm(stdout, rosenbrock, Active, Tuple{Const{Float64}, Active{Float64}})
; @ /home/cceamgi/repo/rsdg-meetings/2023-07-20-julia-ipu/julia-ipu.jl#==#c30b8552-03f4-4f79-b4c0-7179ead69192 within `diffejulia_rosenbrock_11601_inner_1wrap`
; Function Attrs: alwaysinline nofree
define [1 x { double }] @diffejulia_rosenbrock_11601_inner_1wrap(double %0, double %1, double %2) #1 {
entry:
%thread_ptr = call i8* asm "movq %fs:0, $0", "=r"() #6
%tls_ppgcstack = getelementptr i8, i8* %thread_ptr, i64 -8
%3 = bitcast i8* %tls_ppgcstack to {}****
%tls_pgcstack = load {}***, {}**** %3, align 8
%ptls_field.i2.i = getelementptr inbounds {}**, {}*** %tls_pgcstack, i64 2
%4 = bitcast {}*** %ptls_field.i2.i to i64***
%ptls_load.i34.i = load i64**, i64*** %4, align 8
%5 = getelementptr inbounds i64*, i64** %ptls_load.i34.i, i64 2
%safepoint.i.i = load i64*, i64** %5, align 8
fence syncscope("singlethread") seq_cst
; ┌ @ /home/cceamgi/repo/rsdg-meetings/2023-07-20-julia-ipu/julia-ipu.jl#==#c30b8552-03f4-4f79-b4c0-7179ead69192 within `rosenbrock` @ /home/cceamgi/repo/rsdg-meetings/2023-07-20-julia-ipu/julia-ipu.jl#==#c30b8552-03f4-4f79-b4c0-7179ead69192:1
%6 = load volatile i64, i64* %safepoint.i.i, align 8
fence syncscope("singlethread") seq_cst
; │┌ @ intfuncs.jl:332 within `literal_pow`
; ││┌ @ float.jl:411 within `*`
%7 = fmul double %0, %0
; │└└
; │┌ @ float.jl:410 within `-`
%8 = fsub double %1, %7
; │└
; │┌ @ intfuncs.jl:332 within `literal_pow`
; ││┌ @ float.jl:411 within `*`
%9 = fmul fast double %2, 2.000000e+02
%10 = fmul fast double %9, %8
fence syncscope("singlethread") seq_cst
%.unpack1 = insertvalue { double } zeroinitializer, double %10, 0
%11 = insertvalue [1 x { double }] zeroinitializer, { double } %.unpack1, 0
ret [1 x { double }] %11
; └└└
}
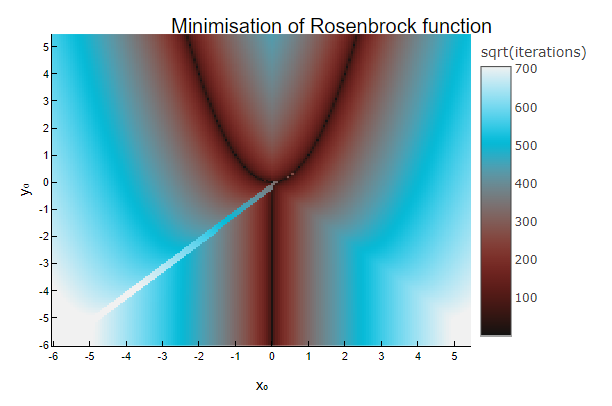
We can run an IPU program to find the minima of the Rosenbrock function on a large grid of different starting points, and plot the number of iterations taken before stopping, when the terminating criteria are met. In particular, we will use the Adam, an optimisation method which requires the gradient of the function to optimise as input, and we will use Enzyme to automatically compute it on the host system at compile time.
rosenbrock(x, y) = (1 - x) ^ 2 + 100 * (y - x ^ 2) ^ 2;
∂(f, x, y) = first(autodiff_deferred(Reverse, f, Active(x), Active(y)));
rosenbrock′(x, y) = ∂(rosenbrock, x, y);
function adam(∂f, x₀::T, y₀::T) where {T}
x, y = x₀, y₀
# Some constants
α = T(0.001) # learning rate
β₁ = T(0.9)
β₂ = T(0.999)
ϵ = T(1e-8)
# Momenta
m = zero(T), zero(T)
v = zero(T), zero(T)
# Stopping criteria
Δ = 10 * eps(T)
δ_x, δ_y = one(T), one(T)
max_t = Int32(500_000)
t = one(max_t)
while abs(δ_x) > Δ < abs(δ_y) && t ≤ max_t
g = ∂f(x, y)
m = β₁ .* m .+ (1 - β₂) .* g
v = β₂ .* v .+ (1 - β₂) .* g .^ 2
m̂ = m ./ (1 - β₁ ^ t)
v̂ = v ./ (1 - β₂ ^ t)
δ_x, δ_y = α .* m̂ ./ (.√(v̂) .+ ϵ)
x -= δ_x
y -= δ_y
t += one(t)
end
# Subtract one because at the end of the last iteration we
# added 1 to the counter but didn't run the following iteration.
return t - one(t)
end;
begin
make_grid(x::T, y::T) where {T<:AbstractVector} =
repeat(x; inner=length(y)), repeat(y; outer=length(x))
# High resolution range
x_range = y_range = collect(Float32.(range(-6; step=2^-4, length=23 * 8)))
# # Low resolution range
# x_range = collect(Float32.(range(-6; step=2^-2, length=23 * 2)))
# y_range = collect(Float32.(range(-8; step=2^-2, length=64)))
end;
m = let
# Inputs to the codelet
x₀, y₀ = make_grid(x_range, y_range)
# Output of the codelet
num_iterations = similar(x₀, Int32)
device = Poplar.get_ipu_device()
target = Poplar.DeviceGetTarget(device)
graph = Poplar.Graph(target)
num_tiles = Int(Poplar.TargetGetNumTiles(target))
@codelet graph function RosenAdam(x::VertexVector{Float32, In},
y::VertexVector{Float32, In},
num_iterations::VertexVector{Int32, Out})
for idx in eachindex(num_iterations)
num_iterations[idx] = adam(rosenbrock′, x[idx], y[idx])
end
end
x₀_ipu = Poplar.GraphAddConstant(graph, x₀)
y₀_ipu = Poplar.GraphAddConstant(graph, y₀)
num_iterations_ipu = similar(graph, num_iterations, Int32);
prog = Poplar.ProgramSequence()
add_vertex(graph, prog, 0:(num_tiles - 1), RosenAdam, x₀_ipu, y₀_ipu, num_iterations_ipu)
Poplar.GraphCreateHostRead(graph, "iterations-read", num_iterations_ipu)
engine = Poplar.Engine(graph, prog)
elapsed = @elapsed Poplar.EngineLoadAndRun(engine, device)
Poplar.EngineReadTensor(engine, "iterations-read", num_iterations)
Poplar.detach_devices()
m = reshape(sqrt.(Int.(num_iterations)), length(y_range), length(x_range))
@info "Loading and running the program on $(length(x₀)) points took $(round(elapsed; sigdigits=4)) seconds"
m
end;
Trying to attach to device 0...
Successfully attached to device 0
Loading and running the program on 33856 points took 30.06 seconds
; ModuleID = 'start'
source_filename = "start"
target datalayout = "e-m:e-p270:32:32-p271:32:32-p272:64:64-i64:64-f80:128-n8:16:32:64-S128"
target triple = "x86_64-linux-gnu"
define void @_Z13_333___Pi_235() local_unnamed_addr #0 !dbg !35 {
top:
%0 = call i64 @get_vec_ptr_Pi(i32 0), !dbg !39
%1 = call i32 @get_vec_size_Pi(i32 0), !dbg !39
%2 = call i64 @get_vec_ptr_Pi(i32 1), !dbg !39
%3 = call i32 @get_vec_size_Pi(i32 1), !dbg !39
%4 = call i64 @get_vec_ptr_Pi(i32 2), !dbg !39
%5 = call i32 @get_vec_size_Pi(i32 2), !dbg !39
%6 = call i32 @llvm.colossus.get.scount.l(), !dbg !40
%coercion = inttoptr i64 %0 to i32*, !dbg !49
%pointerref = load i32, i32* %coercion, align 1, !dbg !49, !tbaa !61, !alias.scope !65, !noalias !68
%7 = call fastcc float @julia_pi_kernel_5874(i32 zeroext %pointerref), !dbg !60
%coercion1 = inttoptr i64 %2 to float*, !dbg !73
store float %7, float* %coercion1, align 1, !dbg !73, !tbaa !61, !alias.scope !65, !noalias !68
%8 = call i32 @llvm.colossus.get.scount.l(), !dbg !81
%9 = sub i32 %8, %6, !dbg !83
%coercion2 = inttoptr i64 %4 to i32*, !dbg !87
store i32 %9, i32* %coercion2, align 1, !dbg !87, !tbaa !61, !alias.scope !65, !noalias !68
ret void, !dbg !91
}
declare i64 @get_vec_ptr_Pi(i32) local_unnamed_addr
declare i32 @get_vec_size_Pi(i32) local_unnamed_addr
declare i32 @llvm.colossus.get.scount.l() local_unnamed_addr
define internal fastcc float @julia_pi_kernel_5874(i32 zeroext %0) unnamed_addr #0 !dbg !92 {
top:
%1 = mul i32 %0, 2917776, !dbg !93
%.not16 = icmp ult i32 %1, -2917775, !dbg !96
%value_phi.v = select i1 %.not16, i32 2917775, i32 -1, !dbg !101
%value_phi = add i32 %1, %value_phi.v, !dbg !101
%.not = icmp ugt i32 %1, %value_phi, !dbg !111
br i1 %.not, label %L78, label %L56, !dbg !95
L56: ; preds = %L56, %top
%value_phi4 = phi i32 [ %9, %L56 ], [ %1, %top ]
%value_phi6 = phi float [ %8, %L56 ], [ 0.000000e+00, %top ]
%2 = uitofp i32 %value_phi4 to float, !dbg !118
%3 = fadd float %2, 0xBFA99999A0000000, !dbg !132
%4 = fmul float %3, 0x3DF0000040000000, !dbg !134
%5 = fmul float %4, %4, !dbg !136
%6 = fadd float %5, 1.000000e+00, !dbg !141
%7 = fdiv float 4.000000e+00, %6, !dbg !144
%8 = fadd float %value_phi6, %7, !dbg !148
%.not15 = icmp eq i32 %value_phi4, %value_phi, !dbg !149
%9 = add i32 %value_phi4, 1, !dbg !151
br i1 %.not15, label %L78, label %L56, !dbg !152
L78: ; preds = %L56, %top
%value_phi10 = phi float [ 0.000000e+00, %top ], [ %8, %L56 ]
ret float %value_phi10, !dbg !153
}
attributes #0 = { "probe-stack"="inline-asm" }
!llvm.module.flags = !{!0, !1}
!llvm.dbg.cu = !{!2, !4, !5, !6, !7, !8, !9, !10, !11, !12, !13, !14, !15, !16, !17, !18, !19, !20, !21, !22, !23, !24, !25, !26, !27, !28, !29, !30, !31, !32, !33}
!julia.kernel = !{!34}
!0 = !{i32 2, !"Dwarf Version", i32 4}
!1 = !{i32 2, !"Debug Info Version", i32 3}
!2 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!3 = !DIFile(filename: "julia", directory: ".")
!4 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!5 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!6 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!7 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!8 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!9 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!10 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!11 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!12 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!13 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!14 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!15 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!16 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!17 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!18 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!19 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!20 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!21 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!22 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!23 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!24 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!25 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!26 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!27 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!28 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!29 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!30 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!31 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!32 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!33 = distinct !DICompileUnit(language: DW_LANG_Julia, file: !3, producer: "julia", isOptimized: true, runtimeVersion: 0, emissionKind: LineTablesOnly, nameTableKind: None)
!34 = !{void ()* @_Z13_333___Pi_235}
!35 = distinct !DISubprogram(name: "#333###Pi#235", linkageName: "julia_#333###Pi#235_5871", scope: null, file: !36, line: 68, type: !37, scopeLine: 68, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!36 = !DIFile(filename: "/home/cceamgi/.julia/packages/IPUToolkit/1u83e/src/compiler/codelet.jl#@#==#892eee7a-73be-4479-8d00-6e1ee8bdd97b", directory: ".")
!37 = !DISubroutineType(types: !38)
!38 = !{}
!39 = !DILocation(line: 69, scope: !35)
!40 = !DILocation(line: 45, scope: !41, inlinedAt: !43)
!41 = distinct !DISubprogram(name: "get_scount_l;", linkageName: "get_scount_l", scope: !42, file: !42, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!42 = !DIFile(filename: "/home/cceamgi/.julia/packages/IPUToolkit/1u83e/src/compiler/runtime.jl", directory: ".")
!43 = !DILocation(line: 100, scope: !44, inlinedAt: !46)
!44 = distinct !DISubprogram(name: "macro expansion;", linkageName: "macro expansion", scope: !45, file: !45, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!45 = !DIFile(filename: "/home/cceamgi/.julia/packages/IPUToolkit/1u83e/src/compiler/timing.jl#@#==#892eee7a-73be-4479-8d00-6e1ee8bdd97b", directory: ".")
!46 = !DILocation(line: 35, scope: !47, inlinedAt: !39)
!47 = distinct !DISubprogram(name: "Pi;", linkageName: "Pi", scope: !48, file: !48, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!48 = !DIFile(filename: "/home/cceamgi/repo/rsdg-meetings/2023-07-20-julia-ipu/julia-ipu.jl#==#892eee7a-73be-4479-8d00-6e1ee8bdd97b", directory: ".")
!49 = !DILocation(line: 119, scope: !50, inlinedAt: !52)
!50 = distinct !DISubprogram(name: "unsafe_load;", linkageName: "unsafe_load", scope: !51, file: !51, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!51 = !DIFile(filename: "pointer.jl", directory: ".")
!52 = !DILocation(line: 46, scope: !53, inlinedAt: !55)
!53 = distinct !DISubprogram(name: "getindex;", linkageName: "getindex", scope: !54, file: !54, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!54 = !DIFile(filename: "/home/cceamgi/.julia/packages/IPUToolkit/1u83e/src/compiler/vertices.jl", directory: ".")
!55 = !DILocation(line: 1336, scope: !56, inlinedAt: !58)
!56 = distinct !DISubprogram(name: "_getindex;", linkageName: "_getindex", scope: !57, file: !57, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!57 = !DIFile(filename: "abstractarray.jl", directory: ".")
!58 = !DILocation(line: 1286, scope: !59, inlinedAt: !60)
!59 = distinct !DISubprogram(name: "getindex;", linkageName: "getindex", scope: !57, file: !57, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!60 = !DILocation(line: 101, scope: !44, inlinedAt: !46)
!61 = !{!62, !62, i64 0}
!62 = !{!"jtbaa_data", !63, i64 0}
!63 = !{!"jtbaa", !64, i64 0}
!64 = !{!"jtbaa"}
!65 = !{!66}
!66 = !{!"jnoalias_data", !67}
!67 = !{!"jnoalias"}
!68 = !{!69, !70, !71, !72}
!69 = !{!"jnoalias_gcframe", !67}
!70 = !{!"jnoalias_stack", !67}
!71 = !{!"jnoalias_typemd", !67}
!72 = !{!"jnoalias_const", !67}
!73 = !DILocation(line: 146, scope: !74, inlinedAt: !75)
!74 = distinct !DISubprogram(name: "unsafe_store!;", linkageName: "unsafe_store!", scope: !51, file: !51, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!75 = !DILocation(line: 42, scope: !76, inlinedAt: !77)
!76 = distinct !DISubprogram(name: "setindex!;", linkageName: "setindex!", scope: !54, file: !54, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!77 = !DILocation(line: 1419, scope: !78, inlinedAt: !79)
!78 = distinct !DISubprogram(name: "_setindex!;", linkageName: "_setindex!", scope: !57, file: !57, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!79 = !DILocation(line: 1389, scope: !80, inlinedAt: !60)
!80 = distinct !DISubprogram(name: "setindex!;", linkageName: "setindex!", scope: !57, file: !57, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!81 = !DILocation(line: 45, scope: !41, inlinedAt: !82)
!82 = !DILocation(line: 102, scope: !44, inlinedAt: !46)
!83 = !DILocation(line: 86, scope: !84, inlinedAt: !86)
!84 = distinct !DISubprogram(name: "-;", linkageName: "-", scope: !85, file: !85, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !2, retainedNodes: !38)
!85 = !DIFile(filename: "int.jl", directory: ".")
!86 = !DILocation(line: 103, scope: !44, inlinedAt: !46)
!87 = !DILocation(line: 146, scope: !74, inlinedAt: !88)
!88 = !DILocation(line: 42, scope: !76, inlinedAt: !89)
!89 = !DILocation(line: 1419, scope: !78, inlinedAt: !90)
!90 = !DILocation(line: 1389, scope: !80, inlinedAt: !46)
!91 = !DILocation(line: 70, scope: !35)
!92 = distinct !DISubprogram(name: "pi_kernel", linkageName: "julia_pi_kernel_5874", scope: null, file: !48, line: 21, type: !37, scopeLine: 21, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!93 = !DILocation(line: 88, scope: !94, inlinedAt: !95)
!94 = distinct !DISubprogram(name: "*;", linkageName: "*", scope: !85, file: !85, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!95 = !DILocation(line: 23, scope: !92)
!96 = !DILocation(line: 513, scope: !97, inlinedAt: !98)
!97 = distinct !DISubprogram(name: "<;", linkageName: "<", scope: !85, file: !85, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!98 = !DILocation(line: 376, scope: !99, inlinedAt: !101)
!99 = distinct !DISubprogram(name: ">;", linkageName: ">", scope: !100, file: !100, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!100 = !DIFile(filename: "operators.jl", directory: ".")
!101 = !DILocation(line: 343, scope: !102, inlinedAt: !104)
!102 = distinct !DISubprogram(name: "steprange_last;", linkageName: "steprange_last", scope: !103, file: !103, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!103 = !DIFile(filename: "range.jl", directory: ".")
!104 = !DILocation(line: 325, scope: !105, inlinedAt: !106)
!105 = distinct !DISubprogram(name: "StepRange;", linkageName: "StepRange", scope: !103, file: !103, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!106 = !DILocation(line: 379, scope: !105, inlinedAt: !107)
!107 = !DILocation(line: 24, scope: !108, inlinedAt: !109)
!108 = distinct !DISubprogram(name: "_colon;", linkageName: "_colon", scope: !103, file: !103, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!109 = !DILocation(line: 22, scope: !110, inlinedAt: !95)
!110 = distinct !DISubprogram(name: "Colon;", linkageName: "Colon", scope: !103, file: !103, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!111 = !DILocation(line: 38, scope: !112, inlinedAt: !114)
!112 = distinct !DISubprogram(name: "&;", linkageName: "&", scope: !113, file: !113, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!113 = !DIFile(filename: "bool.jl", directory: ".")
!114 = !DILocation(line: 669, scope: !115, inlinedAt: !116)
!115 = distinct !DISubprogram(name: "isempty;", linkageName: "isempty", scope: !103, file: !103, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!116 = !DILocation(line: 897, scope: !117, inlinedAt: !95)
!117 = distinct !DISubprogram(name: "iterate;", linkageName: "iterate", scope: !103, file: !103, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!118 = !DILocation(line: 165, scope: !119, inlinedAt: !121)
!119 = distinct !DISubprogram(name: "Float32;", linkageName: "Float32", scope: !120, file: !120, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!120 = !DIFile(filename: "float.jl", directory: ".")
!121 = !DILocation(line: 7, scope: !122, inlinedAt: !124)
!122 = distinct !DISubprogram(name: "convert;", linkageName: "convert", scope: !123, file: !123, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!123 = !DIFile(filename: "number.jl", directory: ".")
!124 = !DILocation(line: 370, scope: !125, inlinedAt: !127)
!125 = distinct !DISubprogram(name: "_promote;", linkageName: "_promote", scope: !126, file: !126, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!126 = !DIFile(filename: "promotion.jl", directory: ".")
!127 = !DILocation(line: 393, scope: !128, inlinedAt: !129)
!128 = distinct !DISubprogram(name: "promote;", linkageName: "promote", scope: !126, file: !126, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!129 = !DILocation(line: 422, scope: !130, inlinedAt: !131)
!130 = distinct !DISubprogram(name: "+;", linkageName: "+", scope: !126, file: !126, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!131 = !DILocation(line: 25, scope: !92)
!132 = !DILocation(line: 410, scope: !133, inlinedAt: !131)
!133 = distinct !DISubprogram(name: "-;", linkageName: "-", scope: !120, file: !120, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!134 = !DILocation(line: 411, scope: !135, inlinedAt: !131)
!135 = distinct !DISubprogram(name: "*;", linkageName: "*", scope: !120, file: !120, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!136 = !DILocation(line: 411, scope: !135, inlinedAt: !137)
!137 = !DILocation(line: 332, scope: !138, inlinedAt: !140)
!138 = distinct !DISubprogram(name: "literal_pow;", linkageName: "literal_pow", scope: !139, file: !139, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!139 = !DIFile(filename: "intfuncs.jl", directory: ".")
!140 = !DILocation(line: 26, scope: !92)
!141 = !DILocation(line: 409, scope: !142, inlinedAt: !143)
!142 = distinct !DISubprogram(name: "+;", linkageName: "+", scope: !120, file: !120, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!143 = !DILocation(line: 422, scope: !130, inlinedAt: !140)
!144 = !DILocation(line: 412, scope: !145, inlinedAt: !146)
!145 = distinct !DISubprogram(name: "/;", linkageName: "/", scope: !120, file: !120, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!146 = !DILocation(line: 425, scope: !147, inlinedAt: !140)
!147 = distinct !DISubprogram(name: "/;", linkageName: "/", scope: !126, file: !126, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!148 = !DILocation(line: 409, scope: !142, inlinedAt: !140)
!149 = !DILocation(line: 521, scope: !150, inlinedAt: !151)
!150 = distinct !DISubprogram(name: "==;", linkageName: "==", scope: !126, file: !126, type: !37, spFlags: DISPFlagDefinition | DISPFlagOptimized, unit: !4, retainedNodes: !38)
!151 = !DILocation(line: 901, scope: !117, inlinedAt: !152)
!152 = !DILocation(line: 28, scope: !92)
!153 = !DILocation(line: 29, scope: !92)
Conclusions 🎉
GPUArrays.jl ➡ explore other programming models (KernelAbstracts.jl, GPUArrays.jl?)Head to https://github.com/JuliaIPU/JuliaIpuDemo and follow the instructions in the README.md for how to play with Julia on the IPU in the cloud with Paperspace for free.
The work of integrating Julia and the IPU was originally started by Emily Dietrich and Luk Burchard at Simula, I recently picked it up and expanded it.
My work was funded by UCL Centre for Advanced Research Computing. I'd like to thank my colleague Sofía Miñano for fruitful discussions.
I'd also like to thank developers and users of GPUCompiler.jl (Valentin Churavy, Tim "maleadt" Besard, Gabriel Baraldi, Valentin "Sukera" Bogad) for their patience in answering my questions, and Graphcore engineers (Mark Pupilli, Andrew Fitzgibbon, Dave Bozier) for their support along the way, and Simula eX³ for the use of their facilities.
Sign up for Graphcore updates:
Apr 28, 2025 \ Research, Scientific Research, LLM

Mar 06, 2025 \ Research, Large models, LLM

Feb 04, 2025 \ AI, Research, LLM

Sign up below to get the latest news and updates: